Segmentation and quantification of subcellular structures in fluorescence microscopy images using Squassh
Type
Programming Language
Interaction Level
License/Openness
Description
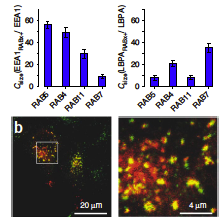
A workflow template to analyze subcellular structures in fluorescence 2D/3D microscopy images based on a Fiji plugin **Squassh** is described in Rizek et al (2014).
The workflow employs detecting, segmenting, and quantifying subcellular structures. For segmentation, it accounts for the microscope optics and for uneven image background. Further analyses include both colocalization and shape analyses. However, it does not work directly for time-lapse data. A brief summary note can be found here.
has topic
has biological terms
Additional keywords
Entry Curator
Post date
12/09/2014 - 15:35
Last modified
05/24/2023 - 19:02